
Home

Upload

Results

Help

Cookies
- {{ channel }} ({{ stdText(info.channel_type) }}): {{ stdChannelInfo(json_data[well], channel) }}
Current COVID19 labs typically have the following procedure:
- 1. Acceptance of samples
- 2. Sample follow-up (LIMS etc.), logistics
- 3. qPCR testing
- 4. Data analysis with machine software and record the result in spreadsheet
- 5. Submit the results to some kind of government registry
Upon discussions, it seems like every step in this process
craves for further optimization. With the genomize-covid19-qPCR
software what we did targets the 4th step above. This step is
error prone due to the 1) manual interpretation of curve
structures of the qPCR results, where a sigmoid curve is
expected, 2) manual recording of results in to spreadsheets.
The sigmoid call for a curve is not a trivial issue, and especially for single channel kits
this process may result to wrong test results. To addres this issue we used machine learning to train a classifier
which considers several features of the curves deemed sigmoid, unsaturated sigmoid or non-sigmoid
in the wet-laboratory tests performed at
Turkish Health Institutions Federation (TÜSEB) COVID19 testing centers.
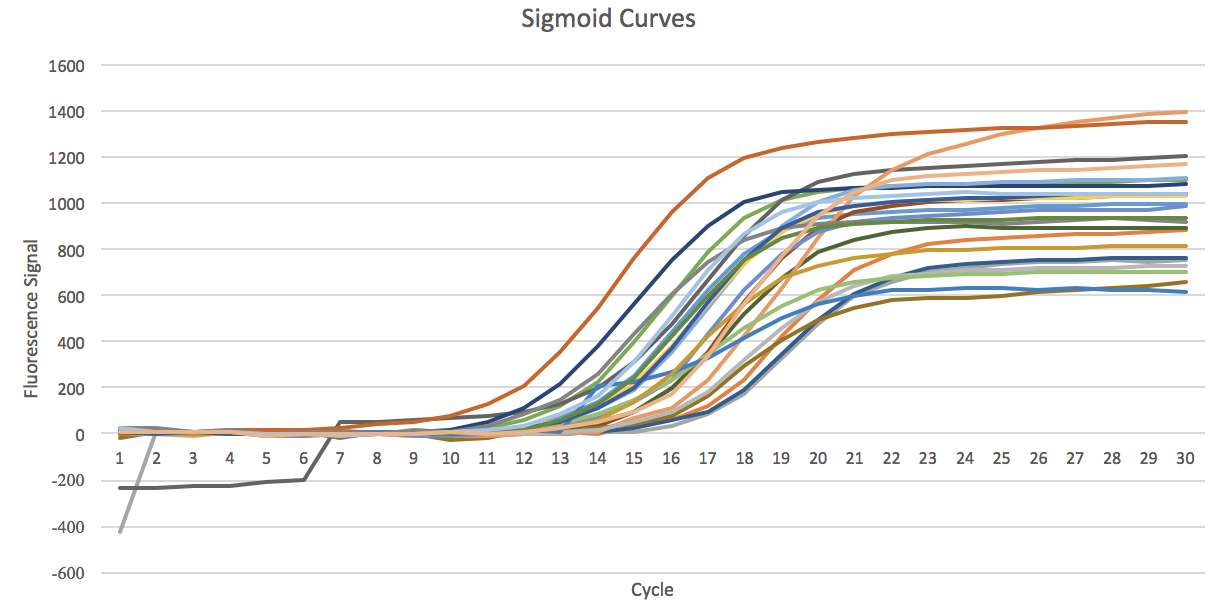
We observed the following type of shapes:
- 1. Fully saturated sigmoid shaped curves, classified as sigmoid
- 2. The curves that has started saturation but not fully saturated, classified as sigmoid
- 3. The curves that has a twisted sigmoid shape, or early accelerating sigmoid curves as we call them, classified as sigmoid
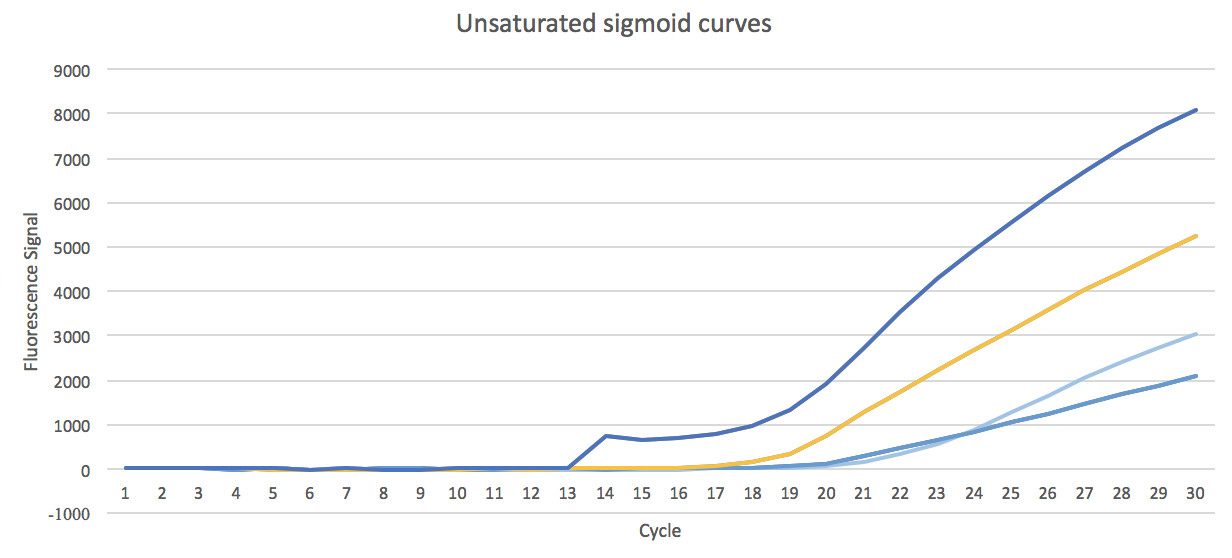
- 4. The curves that did not start saturation, classified as unsaturated sigmoid, and tends to become negative in the retests.
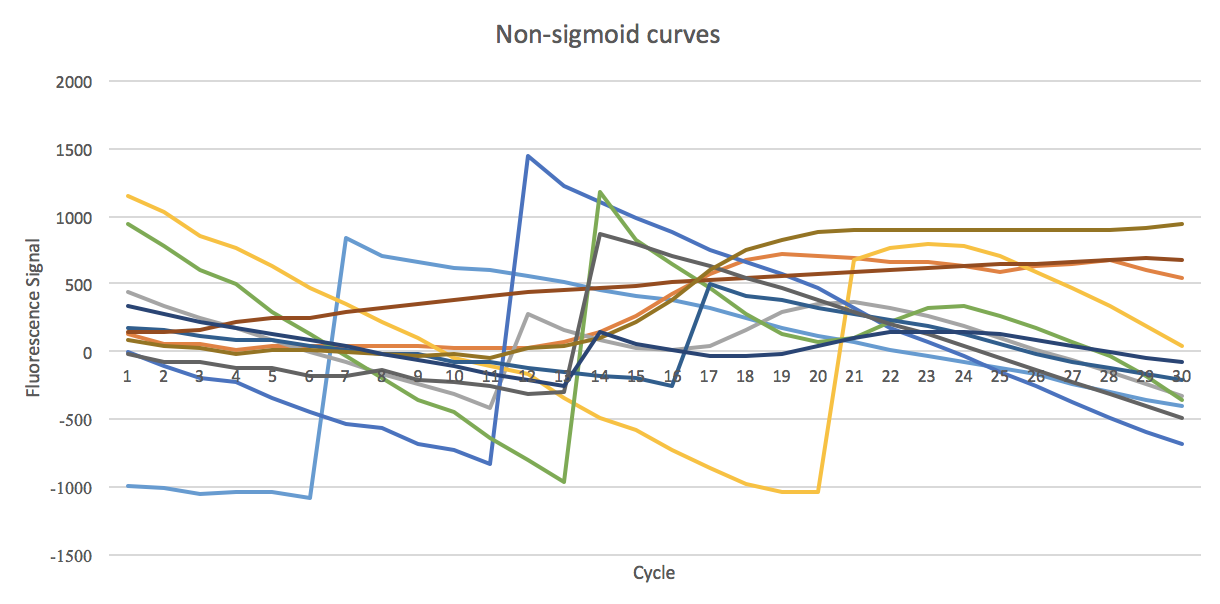
- 5. Non-sigmoid curves, classified as non-sigmoid.
Different Sigmoid Shaped Curves
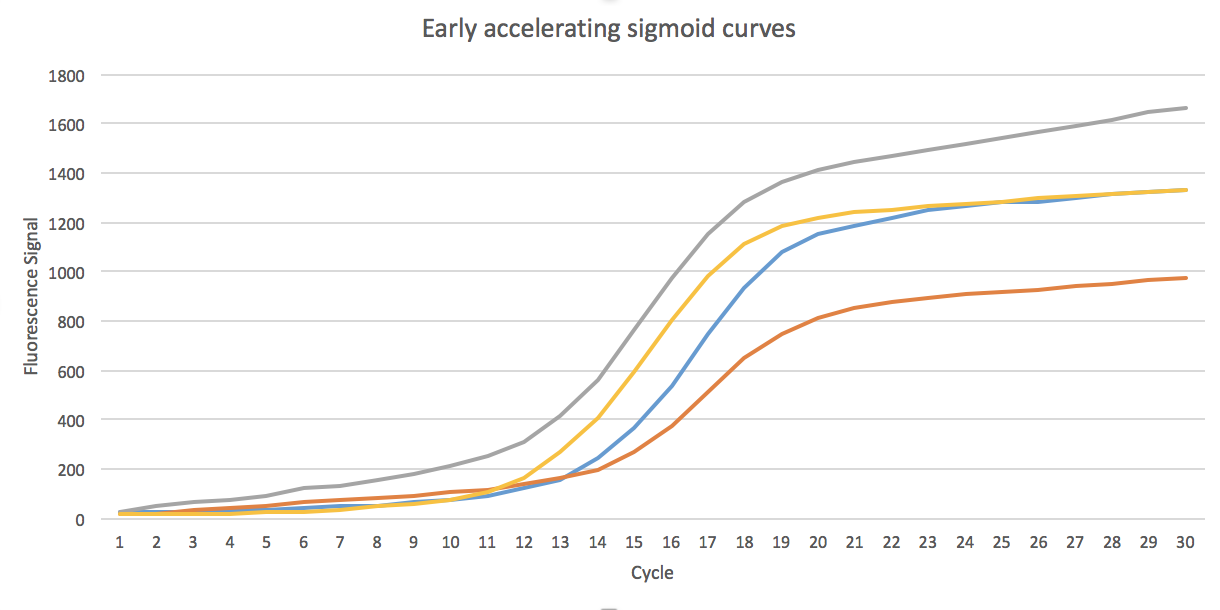
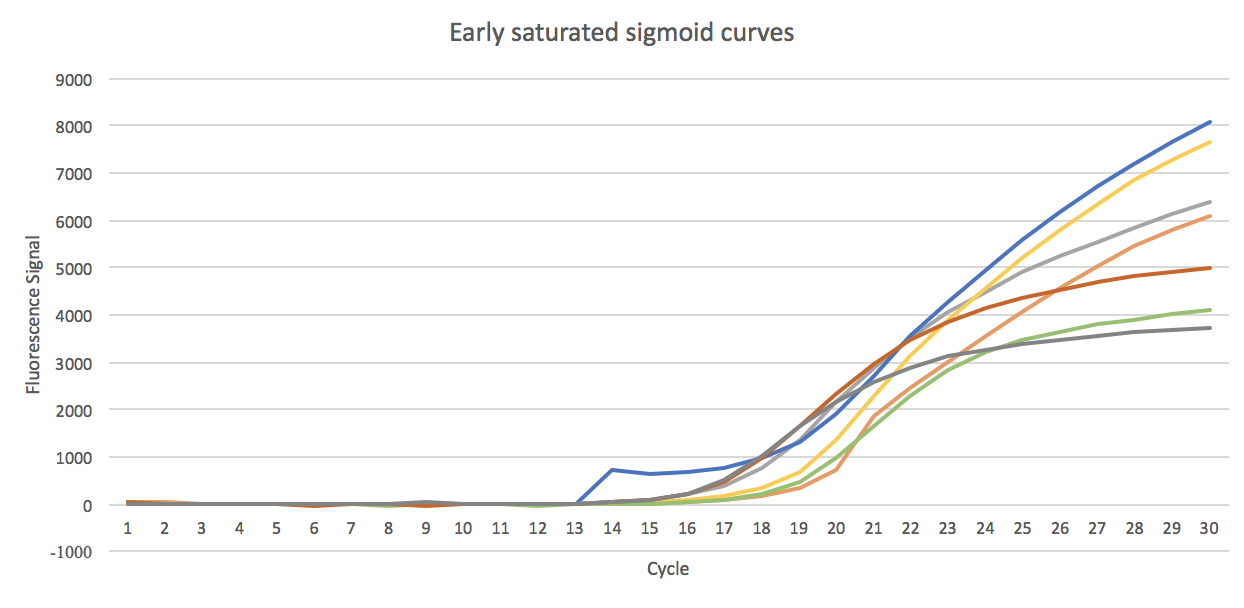
Unsaturated sigmoid shaped curves
Non-sigmoid sigmoid shaped curves
The list of kits defined in the system is below. If the kit you are using is not listed, please send your request to support@genomize.com to have this kit added to the system.
-
{{ kit.name }}
Channels: {{ Object.keys(kit.mutation_list['covid19']).join(', ') }}
Cookies
Selected File:
{{ file.name }} - {{bytesToSize(file.size)}}
| {{ stdText(header) }} |
|---|
| {{ (v) ? stdText(v) : "-" }} |
| {{ stdText(header) }} |
|---|
| {{ (v) ? stdText(v) : "-" }} |
{{ api_error_message }}